
At 37 nM loading concentration, the surface was covered with densely packed ribosomes, often in clear multilayers.

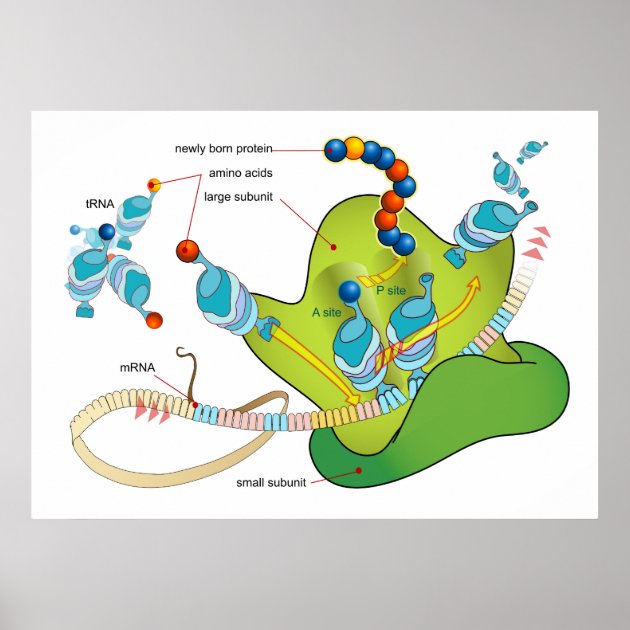
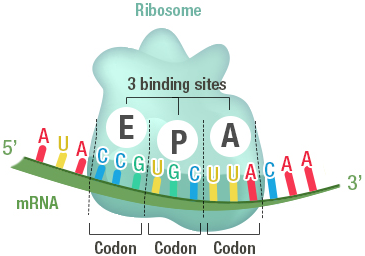
1 ▶) also showed that the density of tightly bound ribosomes is related to the initial concentration applied to the surface. Surface images of ribosomes adsorbed to mica obtained by tapping-mode atomic force microscopy (Fig. For the experiments described below, the number of ribosomes present in solution due to detachment from the surface was negligible. These measurements indicate that at 500 nM, a multilayer of ribosomes (4–5 deep) is obtained, whereas at 10 nM, a sparse distribution of ribosomes is achieved, with ~8% of the surface occupied. We obtained 1.5 ± 0.2 ( n = 4) and 0.020 ± 0.002 ( n = 21 data not shown) pmole of bound ribosomes/cm 2 for the higher and lower concentrations, respectively. At both loading concentrations, a layer of stably bound ribosomes remained on the surface after extensive washing, although removal of non-stably bound ribosomes required more washes at the lower applied concentration. Quantification of adsorbed material was achieved using 70S particles labeled with -phosphate. These concentrations approximate the conditions used in measuring poly(Phe) synthesis in bulk solution and ribosome activity in the microscopic experiments or poly(Phe) synthesis on mica, respectively. Ribosomes were adsorbed to mica sample chambers from solutions containing 500 nM or 10 nM concentrations of 70S particles. We investigated whether poly(U)-programmed ribosomes, adsorbed nonspecifically to mica surfaces, could synthesize TCA-precipitable poly(-Phe) and translocate along poly(U) mRNA. Here we extend this work by measuring bulk poly(Phe) synthesis on poly(U)-programmed mica-bound ribosomes and detecting translocation of individual ribosome-bound poly(U) templates using optical microscopy to examine mRNA-tethered beads. It has been shown previously that ribosomes adsorbed on a mica surface are competent to bind aminoacyl-tRNA in the peptidyl transfer site (P-site) and to form a peptide bond with added puromycin ( Sytnik et al. For instance, applying an external force to the mRNA and measuring the relationship between the rate of translocation and the force applied could help to distinguish between proposed mechanisms of translocation ( Czworkowski and Moore 1997 Keller and Bustamante 2000 Wintermeyer and Rodnina 2000). Because the sliding of the ribosome along the mRNA template is a mechanical output, analogous single-molecule measurements should provide information relevant toward the understanding of the molecular mechanism of protein synthesis. 2000), helping to elucidate their properties and mechanisms. 1999 Ishijima and Yanagida 2001) and nucleic acid processing enzymes ( Rich 1998 Wang et al. Single-molecule techniques have been used to measure directly the elementary events of production of force and displacement by molecular motors ( Kinosita 1999 Mehta et al. The mechanisms of action of these factors are not fully understood because the structural changes they undergo and the mechanical events they facilitate have not been elucidated on functioning ribosomes. Although the energy required for formation of the peptide bond comes from the ester bond of tRNA charged with its cognate amino acid, the remarkable rate of translation (10–20 peptide bonds/sec Kjeldgaard and Gausing 1974 Kennell and Riezman 1977), fidelity of amino acid selection, and maintenance of the reading frame during translocation (~10 −4 error rate Loftfield and Vanderjagt 1972 Kurland 1992) require the GTPase activities of two G-protein elongation factors, EF-Tu and EF-G. The kinetics of the elongation cycle have been elucidated by steady-state and transient kinetics studies ( Rodnina et al. 2000 Frank 2001) and X-ray diffraction studies ( Ban et al. To understand the role of tRNA, you need to know more about its structure.Astonishing progress on the structure of the ribosome and its accessory factors has been obtained from cryo-electron microscopy ( Stark et al. The ribosome reads the sequence of codons in mRNA, and molecules of tRNA bring amino acids to the ribosome in the correct sequence. Translation happens on the ribosomes floating in the cytosol, or on the ribosomes attached to the rough endoplasmic reticulum. After mRNA leaves the nucleus, it moves to a ribosome, which consists of rRNA and proteins.


 0 kommentar(er)
0 kommentar(er)
